In the bioinformatics research field, NextFlow is a mainstream data analysis tool widely used in many research projects. MemVerge, a US company specializing in memory-convergence infrastructure, has seamlessly integrated NextFlow into its public cloud computing platform product, Memory Machine Cloud (MMCloud). This integration provides bioinformaticians with automated high-performance computing, checkpoint and restore functionalities, and optimized cloud host selection. This enables users to work without needing to adapt to a new and complex infrastructure management environment.
To execute NextFlow pipelines, shared storage was crucial. Our main challenges included:
- The need to handle tasks with different I/O requirements, involving the operation of large files and massive small files.
- POSIX compatibility was essential due to most task scripts supporting file protocols only.
- In a workflow, the execution order of tasks depended on their dependencies, making real-time status crucial.
After we evaluated various storage solutions including Network File System (NFS), s3fs, Amazon Elastic File System (EFS), and FusionFS, we chose JuiceFS, an open-source distributed file system. JuiceFS ensures a high-performance shared storage solution, particularly excelling in scenarios with a large number of small files. In our tests, the average number of small files written per minute was five times that of s3fs. Furthermore, JuiceFS has a lower cost.
In this post, we’ll deep dive into our storage pain points, explain why we chose JuiceFS over NFS, s3fs, EFS, and FusionFS, and share our future optimization plans.
Storage pain points of using NextFlow on the cloud
NextFlow + MemVerge
In the bioinformatics field, NextFlow is a widely used workflow management tool. Originating in Spain, it has the largest user base in the United States, with substantial usage in the UK, Germany, and Spain. NextFlow is renowned for its excellent workflow management capabilities, automating complex compute processes and significantly simplifying operations.
NextFlow does not provide resource management and scheduling features, relying on third-party schedulers to execute each compute task. The caching mechanism is crucial for NextFlow. Many research projects frequently access public data resources stored on remote services such as S3. Directly loading data from S3 each time would significantly reduce efficiency. Therefore, NextFlow uses shared storage provided by schedulers like MemVerge to cache data. This improves data access speed and optimizes overall compute performance.
To further enhance the efficiency of NextFlow, we developed the open-source plugin nf-float. This plugin connects NextFlow with MemVerge's MMCloud, allowing all task scheduling to be managed through the MMCloud OpCenter. In OpCenter, we precisely allocate and use resources in collaboration with cloud providers like AWS and Alibaba Cloud, using cloud-native APIs. New cloud host instances are dynamically configured for each task, with monitoring of task execution and resource usage to ensure optimal efficiency and cost-effectiveness. Learn more about the MemVerge + NextFlow solution.
Storage challenges in using NextFlow
In the architecture of NextFlow, shared storage is a critical mechanism. It not only tracks the execution status of each task to ensure the smooth operation of the entire workflow but also facilitates data sharing. This allows the output of one task to become the input for another and thus accelerates data flow and interaction between tasks. We explored various options for shared storage, and our requirements included:
- Performance: Since shared storage directly affects the execution efficiency of the entire workflow, its performance must meet high standards. Slow storage response times could delay task execution and significantly reduce overall workflow speed, creating bottlenecks.
- Scalability: Given the significant differences in data needs between different workflows and tasks within the same workflow, shared storage must be highly scalable to handle varying data capacity requirements and ensure efficient resource usage.
- POSIX compatibility: Most bioinformatics tasks are designed based on POSIX standard file system interfaces. The developers of these tasks are typically scientists rather than computer scientists. Scientists prefer intuitive file operation methods for data handling. Therefore, the shared storage system needs to seamlessly support POSIX interfaces to lower the technical barrier.
- Cost-effectiveness: This is a crucial factor for research institutions when choosing a storage solution. Especially in cloud computing environments, optimizing storage solutions to enhance cost-effectiveness is a core consideration. Thus, controlling storage costs is particularly important.
Why we chose JuiceFS
EFS vs. s3fs vs. FusionFS vs. JuiceFS
In the initial trial phase, we set up an NFS server as a shared volume but found its performance inadequate for handling large-scale workflows. Then, we switched to AWS EFS, which offered good performance, but its high costs and inconvenient data transfers with object storage limited its suitability.
To balance performance and compatibility, we tried the s3fs solution, which provided a POSIX file system interface. However, its performance significantly dropped when handling a large number of small files and frequent write operations. It couldn’t meet high-performance requirements.
Some users, in pursuit of higher performance, turned to Amazon FSx for Lustre. While Lustre demonstrated excellent performance, its high costs made it unaffordable for most users.
To clearly identify the differences between these solutions, we conducted an RNA-Seq test. RNA-Seq is a technique used to analyze the expression levels of various RNAs in cells. It’s a very complex workflow containing up to 276 tasks.
In this test, we evaluated the time and cost of AWS EFS, s3fs, JuiceFS, and FusionFS. FusionFS is a distributed file system developed by Seqera, the commercial provider of NextFlow, specifically for bioinformatics research. Time is measured using two metrics:
- CPU time, the total cumulative time spent by the CPU.
- Wall time, the total elapsed time from the start to the end of the task.
The following table shows the test results:
| Storage solution | CPU time | Wall time | Cost |
|---|---|---|---|
| EFS with MMCloud | 483.5 | 7h 16m 51s | High |
| s3fs with MMCloud | 1586 hrs | 26 hrs | High |
| FusionFS with AWS Batch | 647.4 | 7h 45m 18s | $90.34 + Fusion License |
| JuiceFS with MMCloud | 485.6 | 6h 56m 29s | $20.84 |
The results show that:
- AWS EFS with MMCloud: The entire workflow was completed in 7 hours, but with high costs.
- s3fs with MMCloud: The time increased significantly to 26 hours. Although s3fs was low-cost, the extended runtime resulted in high overall costs.
- FusionFS with AWS Batch: The workflow was completed in 7 hours and 45 minutes, with cloud resource costs reaching $90, plus additional fees for Seqera.
- JuiceFS with MMCloud: The task was completed in 6 hours and 56 minutes, costing only $20. It was the best-performing solution in this test.
Performance of writing massive small files: JuiceFS vs. s3fs
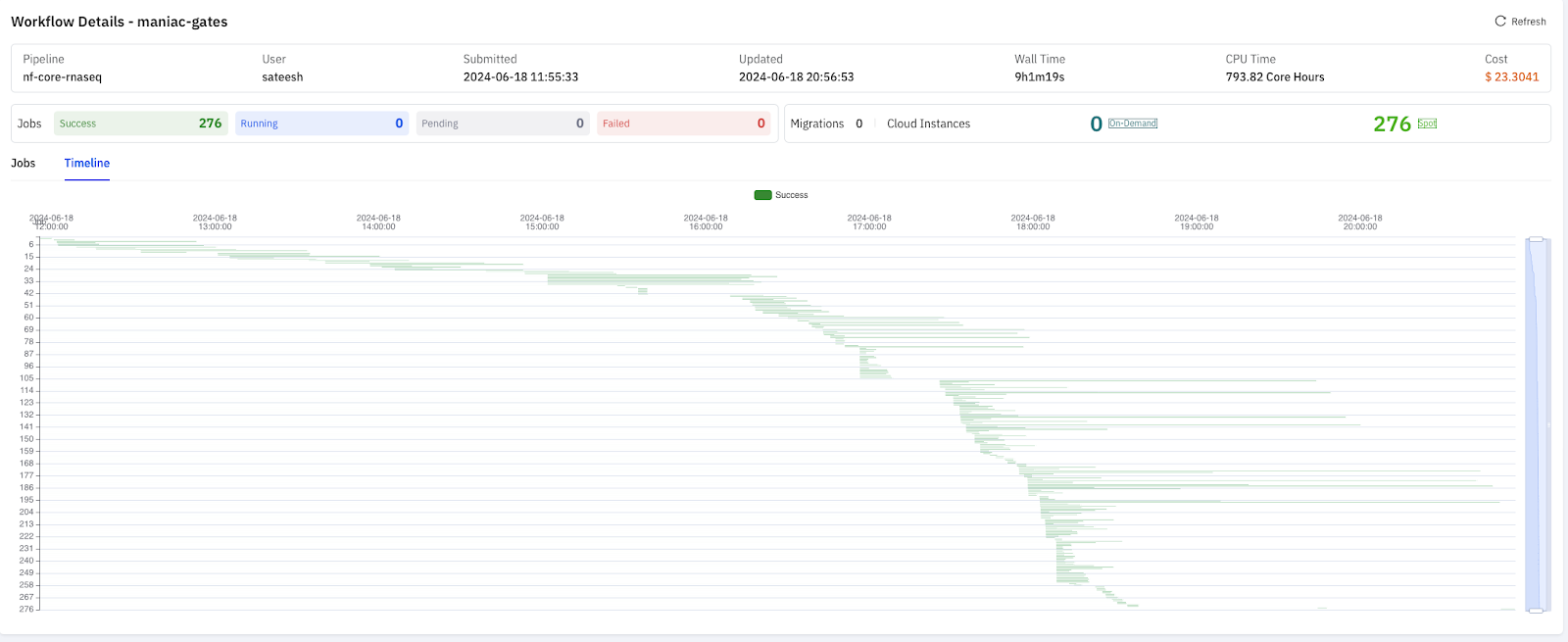
To understand the differences between s3fs and JuiceFS beyond overall runtime, we conducted another RNA-Seq test with over 200 tasks. As shown in the figure below, each line represents a task's execution, and the length indicates the task's duration.
JuiceFS performed well in managing concurrent and interdependent tasks. It completed the entire process in 9 hours at a cost of only $23.
When using s3fs, the workflow failed after running for 9 hours due to several tasks taking excessively long to complete, resulting in a cost of $37.
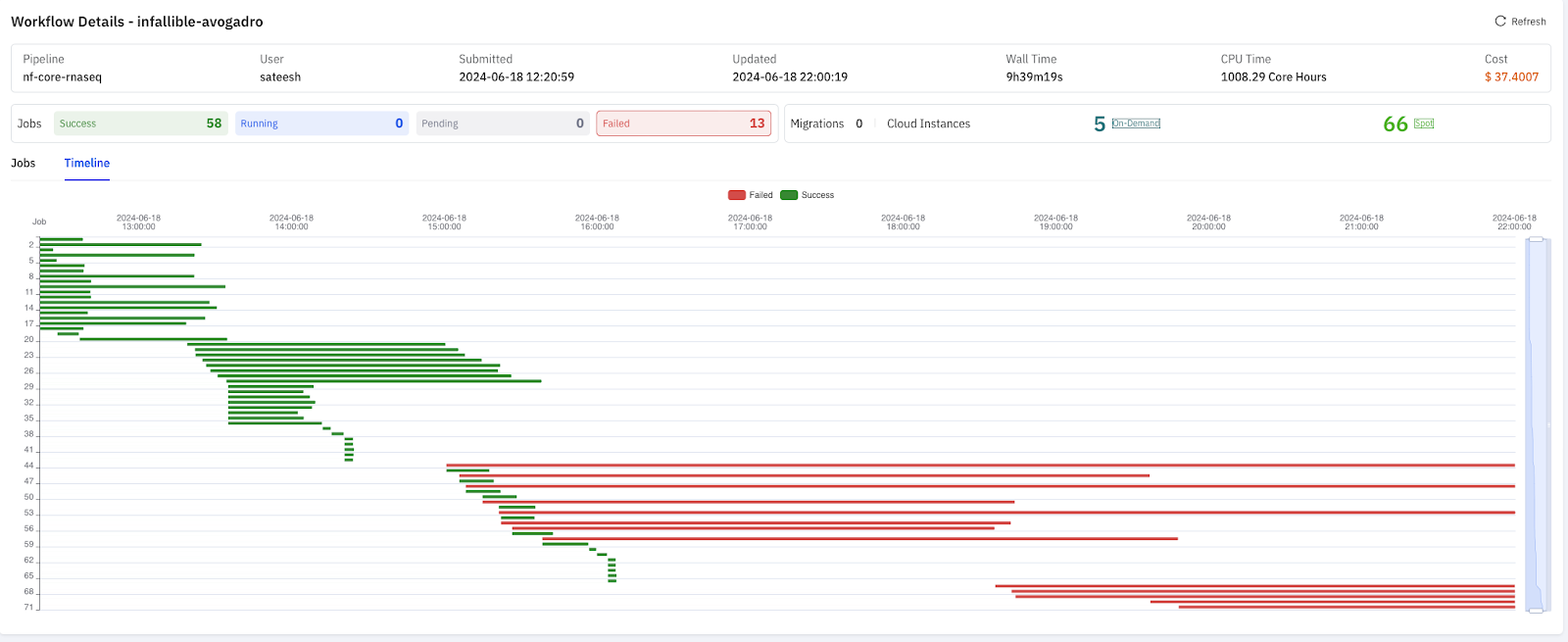
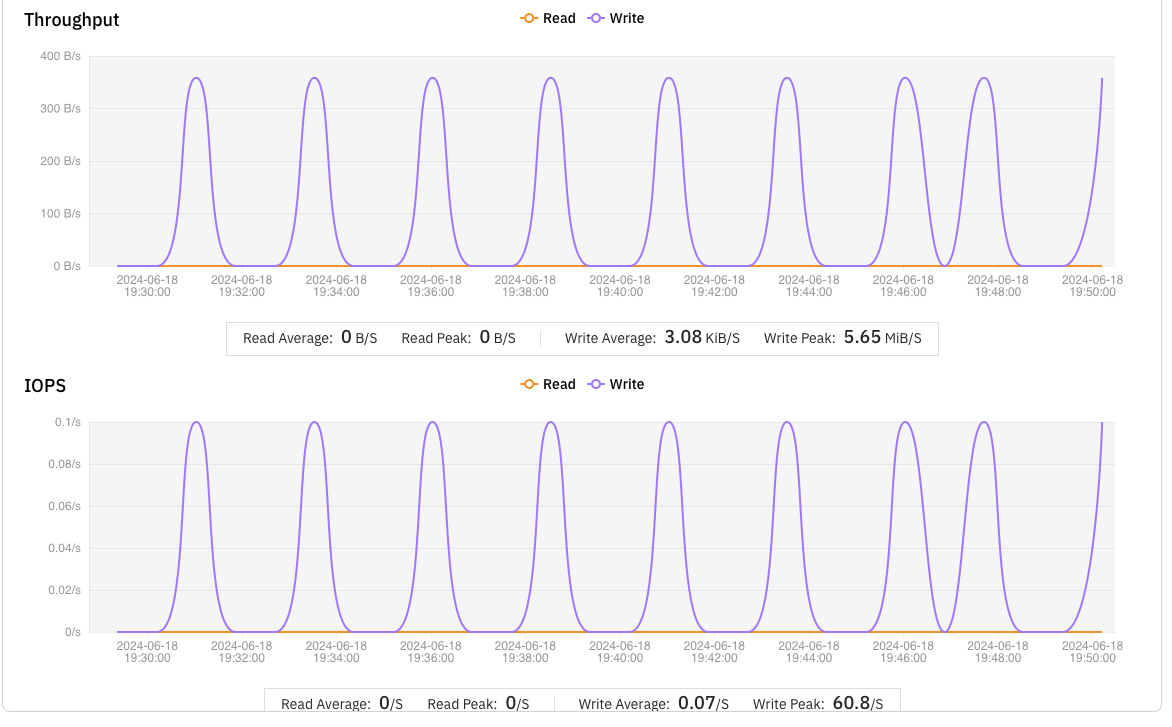
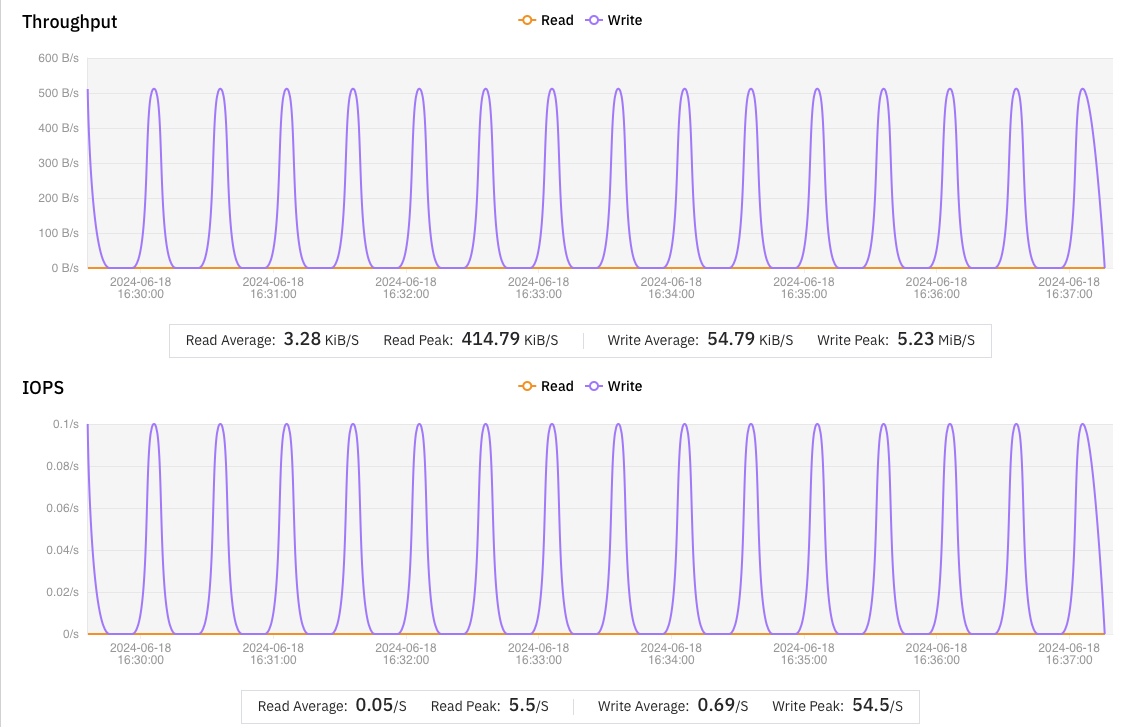
We analyzed the failed tasks with s3fs and found that the issues were due to the frequent writing of small files less than 500 bytes in size.
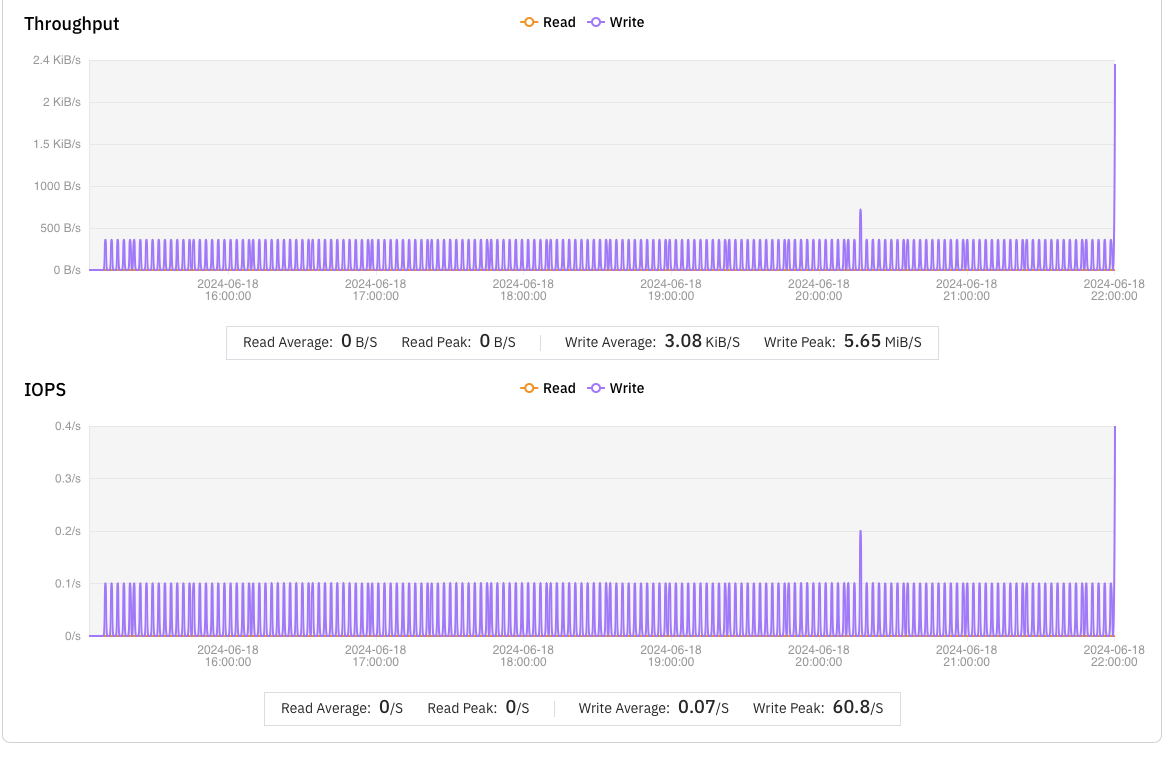
s3fs was inefficient at handling such small files, averaging only 0.5 files per minute. This caused many tasks to take too long and be marked as failed by NextFlow. In contrast, JuiceFS averaged 2.5 small files per minute, performing five times better than s3fs.
JuiceFS showed significant advantages in performance and cost-effectiveness, so it was our final choice.
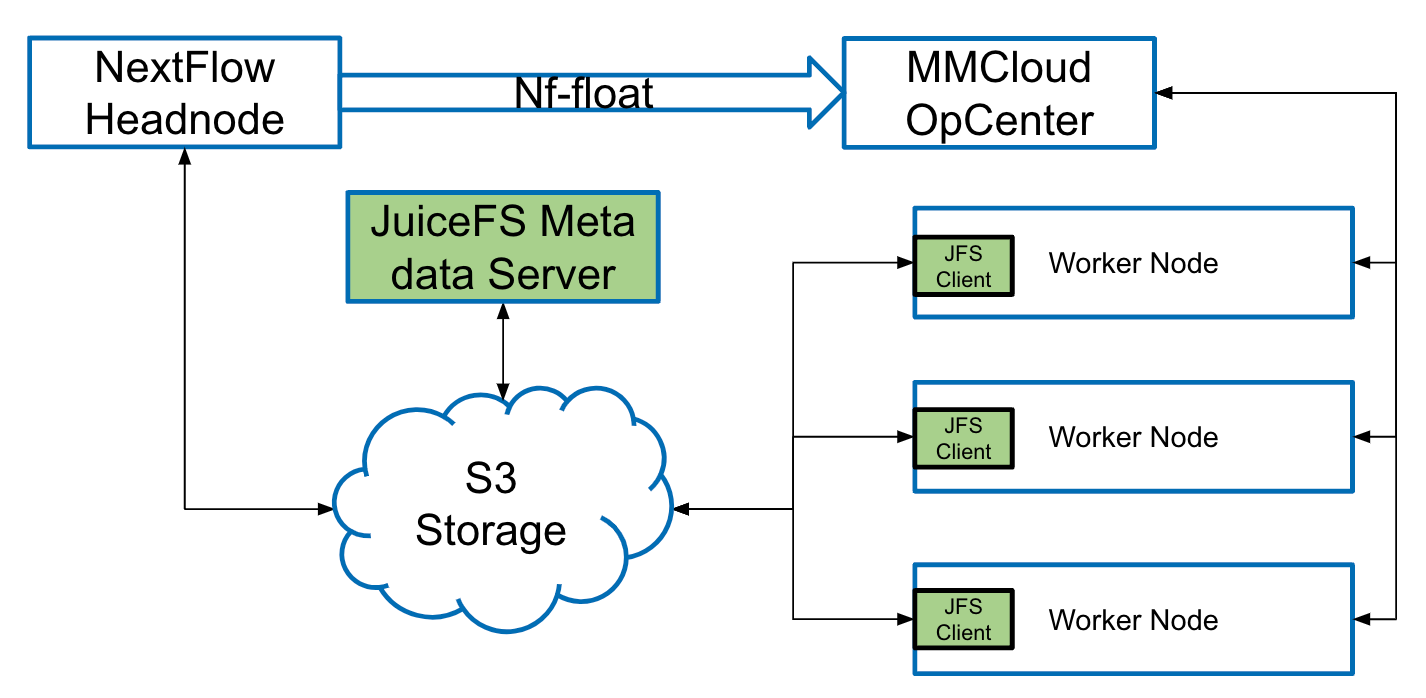
In our architecture, when NextFlow triggers a task, we dynamically create a JuiceFS instance using the OpCenter mechanism. Users need to provide the location of the object storage, which is then shared by OpCenter, the NextFlow Headnode, and all related parties. This fully meets our requirements.
On-demand instances vs. spot instances
Public cloud providers offer two types of instances: on-demand instances and spot instances. In the spot instance mode, MMCloud seeks higher-performing models at the same price, thereby reducing task execution time. Leveraging this feature, we tested JuiceFS under both modes.
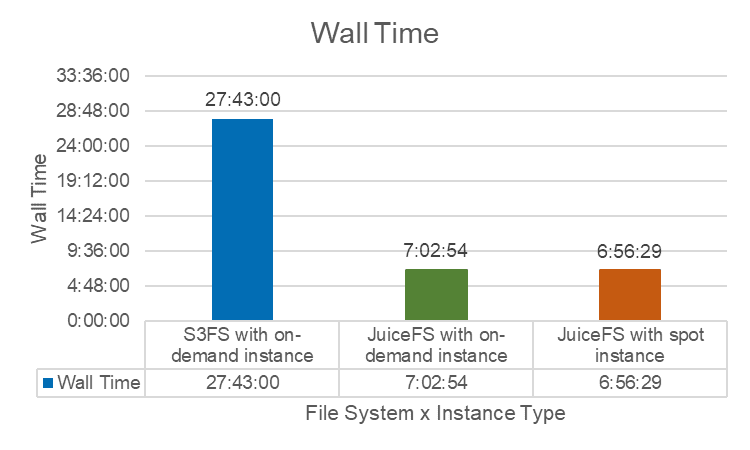
As expected, JuiceFS performed better on MMCloud's spot instances. We also tested s3fs, which took significantly longer than JuiceFS on both instance types.
Future optimization plans for JuiceFS
In the future, we’ll focus on optimizing data preprocessing and transfer, exploring shared cache mechanisms, and fine-tuning JuiceFS parameters to further improve the execution efficiency and performance of NextFlow workflows. Optimizing data preprocessing and transfer
Currently, many users store their raw datasets in object storage services like S3. In NextFlow, if the shared storage is defined with a POSIX interface, the system may automatically copy data from S3 to local shared storage. This process, known as data staging, is often time-consuming and inefficient, because the data is already stored in S3 and doesn't need to be copied.
Therefore, we plan to introduce direct S3 interfaces as the data access layer for NextFlow. This will eliminate unnecessary data staging steps and improve overall workflow efficiency.
Cache group optimization
For JuiceFS Cloud Service and large-scale parallel tasks, we noticed that when a NextFlow workflow launched a large number of tasks simultaneously (for example, 200 tasks), each task may independently cache the same reference data. This led to redundant data transfer and storage, thus wasting compute resources.
To address this issue, we plan to explore a shared cache mechanism, ensuring that all tasks can share the same cached data. This will avoid redundant caching at each node and significantly improve data utilization efficiency and overall performance.
Mount point parameter optimization
In the current JuiceFS mounting process, we use default parameter configurations. This limits the maximum performance of tasks to some extent.
To further enhance the execution efficiency of each task, we plan to fine-tune and optimize JuiceFS parameters based on the specific attributes and requirements of the tasks. By customizing the parameter configurations, we can better match the resource needs and execution characteristics of the tasks, thus maximizing performance.
If you have any questions or would like to learn more, feel free to join JuiceFS discussions on GitHub and their community on Slack.
About MemVerge
MemVerge, based in Silicon Valley, USA, specializes in memory technology development. Our product, MMCloud, uses patented memory management technology to intelligently schedule cloud resources. It significantly enhances the usability and efficiency of cloud computing. Our company has attracted investments from industry leaders such as Intel and NetApp.
One of our primary focus areas is bioinformatics, particularly supporting companies that require large-scale genetic analysis. This field typically demands high-performance computing to process complex big data. Facing the high costs and resource constraints of data center construction, many bioinformatics companies are turning to cloud computing platforms to execute their compute tasks.
MemVerge's star customers include:
- HZPC from the Netherlands, a global leader in potato genetics research.
- oNKo-innate from Australia, a highly acclaimed cancer research pioneer in the medical community.
- MDI BioScience from the United States, a century-old institution dedicated to studying the regenerative capabilities of - - certain lizards, aiming to advance human medicine through genetic analysis.


















